Genomic nucleosome organization reconstituted with pure proteins
News from the Korber group
29.11.2016
Nils Krietenstein, Megha Wal, Shinya Watanabe, Bongsoo Park, Craig L. Peterson, B. Franklin Pugh, Philipp Korber (2016) Genomic Nucleosome Organization Reconstituted with Pure Proteins. Cell 167(3), p709-721.
DOI: http://dx.doi.org/10.1016/j.cell.2016.09.045
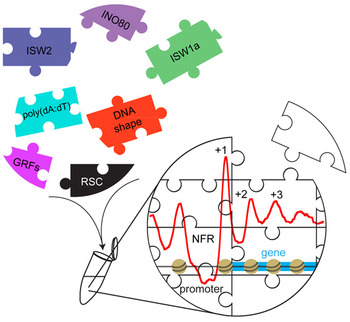 The positioning of nucleosomes along the DNA is a fundamental level of genome regulation for eukaryotes. If a certain DNA sequence is readily accessible for factors that drive, for example, transcription or replication, is critically affected by the presence of absence of nucleosomes. So understanding the molecular mechanisms that determine nucleosome positioning is an important goal. The graduate student Nils Krietenstein in the group of PD Philipp Korber (BMC, LMU Munich), in collaboration with the groups of Prof. B. Franklin Pugh (Pennsylvania State University, USA) and Prof. Craig L. Peterson (University of Massachusetts Medical School, USA), succeeded in reconstituting in vivo-like promoter nucleosome patterns across the budding yeast genome with purified factors (Krietenstein*, Wal*, et al., 2016, Cell). This rather unique genome-wide biochemistry approach allows identifying the direct, specific and to a surprisingly large extent sufficient contribution of factors, especially ATP-dependent nucleosome remodeling enzymes and so called barrier factors. It turns out that these factors generate a dynamic, self-organizing ground-state of nucleosome positioning that other genomic processes work on.
The positioning of nucleosomes along the DNA is a fundamental level of genome regulation for eukaryotes. If a certain DNA sequence is readily accessible for factors that drive, for example, transcription or replication, is critically affected by the presence of absence of nucleosomes. So understanding the molecular mechanisms that determine nucleosome positioning is an important goal. The graduate student Nils Krietenstein in the group of PD Philipp Korber (BMC, LMU Munich), in collaboration with the groups of Prof. B. Franklin Pugh (Pennsylvania State University, USA) and Prof. Craig L. Peterson (University of Massachusetts Medical School, USA), succeeded in reconstituting in vivo-like promoter nucleosome patterns across the budding yeast genome with purified factors (Krietenstein*, Wal*, et al., 2016, Cell). This rather unique genome-wide biochemistry approach allows identifying the direct, specific and to a surprisingly large extent sufficient contribution of factors, especially ATP-dependent nucleosome remodeling enzymes and so called barrier factors. It turns out that these factors generate a dynamic, self-organizing ground-state of nucleosome positioning that other genomic processes work on.
Philipp Korber heads the project A04 in the CRC; Nils Krietenstein is an IRTG 1064 alumnus.
Link to Cell paper:
http://www.cell.com/cell/fulltext/S0092-8674(16)31330-7
See LMU press release http://www.en.uni-muenchen.de/news/newsarchiv/2016/korber_dnaverpackung.html

