A23 - Chromatin as limiting factor in the regulation of DNA repair pathway choice
A stable genome is a fundamental requirement of life. This stability is however constantly challenged by damages to the DNA – a driving force in cancer development. In eukaryotic cells DNA damage does not occur on “naked” DNA, but in the context of chromatin. Therefore, the cellular DNA damage response is adapted to this feature. On the one hand, chromatin constitutes a barrier to repair. On the other hand, chromatin is now increasingly recognized as a crucial regulator of the repair machinery. We have recently shown for the repair of DNA double-strand breaks (DSBs) that chromatin can be in two states, which are either permissive or restrictive to a process called DNA end resection (Bantele et al. 2017, eLIFE). Resection is a crucial process that determines whether a DSB is repaired by homologous recombination or alternative repair pathways such as non-homologous end-joining. Understanding how cells control DNA end resection and thereby pathway choice will therefore not only give fundamental insights into an essential cell biological mechanism, but also have a critical impact on future recombination-dependent gene editing approaches (e.g. CRISPR/Cas9).
We have developed an experimental toolbox (Figure 1), with which we now can shift damaged chromatin into a resection-permissive (Figure 1, left side) or a resection-restrictive state (Figure 1, right side). With this system, we are in a strong position to characterize the changes that occur within chromatin in a systems biology approach, when it switches between the two states (project I).
Furthermore, we are using in vitro reconstitution to reveal the biochemical mechanism, by which resection inhibitors such as budding yeast Rad9 work (Figure 2, left hand side panels for models). Additionally, we aim to reveal how such inhibition is overcome by nucleosome remodellers that promote resection (Figure 2, right hand side panels for models; project II).
Collectively, both projects will allow unprecedented insights into how cells modulate chromatin in order to control chromatin-associated processes.
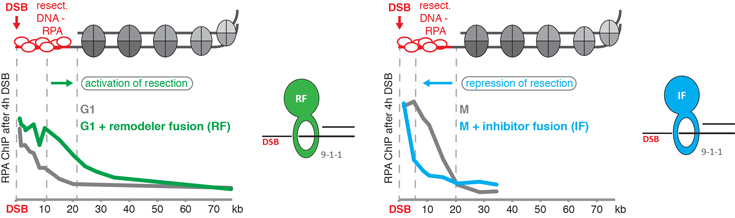
Figure 1: Our experimental toolbox to induce resection-permissive and resection-restrictive chromatin states. ChIP-based analysis of DNA end resection surrounding a site-specific DSB: left: a protein fusion of the 9-1-1 complex to a nucleosome remodeller (RF; green) artificially activates DNA end resection, when it is physiologically inactive (G1 phase); right: a protein fusion of the 9-1-1 complex to a resection inhibitor (IF; blue) artificially blocks DNA end resection, when it is physiologically active (M phase).
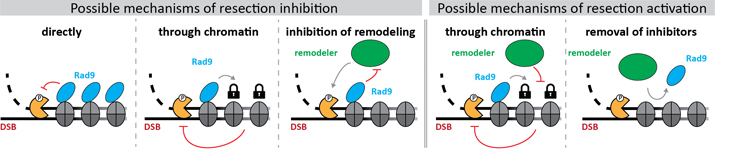
Figure 2: Models for the molecular mechanisms, by which resection inhibitors (blue) block DNA end resection (left) and by which nucleosome remodellers (green) promote DNA end resection (right).

Prof. Dr. Boris PfanderProfessor for Genome Maintenance Mechanisms in Health and Disease, DLR Cologne +49 2203 601 1277 |

