A04 - Genome-wide reconstitution of dynamics in nucleosome positioning, collision, eviction, and histone exchange

We use a unique genome-wide chromatin reconstitution system with purified factors to explore remodeler mechanisms of nucleosome positioning, collision, eviction, generation of subnucleosomal particles and histone exchange, at steady state and during kinetics. The genome-wide approach allows direct comparisons with corresponding in vivo data. With Hopfner (A06) we will include structure-guided yeast INO80 mutant remodelers and human INO80. Probing the role of genomic DNA sequence, barriers, histone modifications/variants and histone chaperones will reveal how remodelers process multiple levels of information into remodeler-specific outcomes.
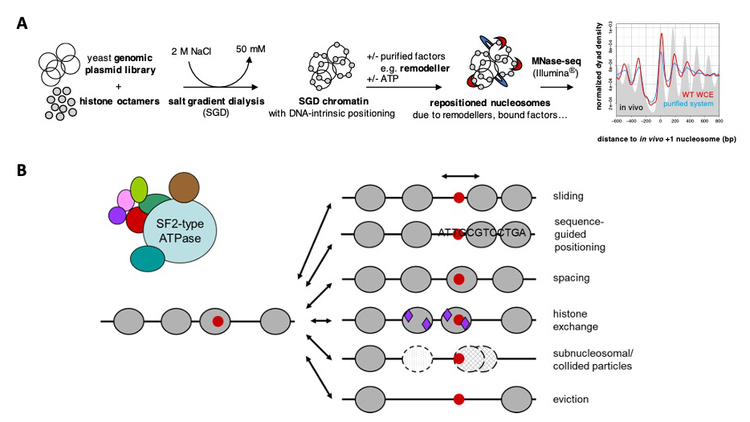
A: Schematic of our genome-wide reconstitution system (Zhang et al., 2011, Science; Krietenstein et al., 2012, Meth. Enzymol.; Krietenstein et al., 2016, Cell; Oberbeckmann, Krietenstein et al., 2021, Nat. Comm.; Oberbeckmann, Niebauer et al., 2021, Nat. Comm.).
B: Overview of ATP-dependent nucleosome remodeling outcomes.


