A17 - Role and Regulation of DNA Modifications
We have previously shown that the multidomain protein UHRF1 controls DNA methylation via two distinct pathways, by dual monoubiquitination of histone H3 or the PCNA associated PAF15, which both activate DNMT1. Interestingly, UHRF1 is inhibited by the recently evolved and TET-controlled DPPA3 which can genome-wide erase DNA methylation. We now want to investigate how the activity of UHRF1 is regulated by posttranslational modifications combining mutagenesis with analyses of DNA methylation and chromatin complexes. These experiments should help to further elucidate the role and regulation of UHRF1 in the dynamic interplay between histone and DNA modifications.
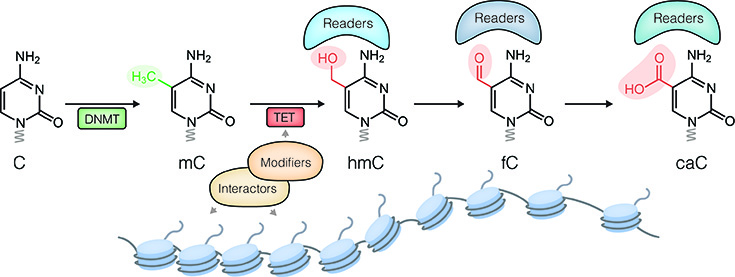
Dynamic changes of DNA modifications and chromatin states: Establishment, maintenance and dynamic changes of functional chromatin states

Prof. Dr. Heinrich LeonhardtHuman Biology & Bioimaging, Faculty of Biology, LMU Munich +49 (0)89 2180 - 74232 |

