A12 - Histone H4K20 demethylation in cell cycle-controlled differentiation
Our current results from the model organism Xenopus indicate that Histone H4K20me1 and -me2 levels are regulated by S-phase dilution in proliferating cells, while becoming adjusted by specific histone demethylases such as PHF8, RSBN1 and HR23B in postmitotic, differentiating cells. By combining gain and loss of function analysis with RNA-Seq and ChIP-Seq approaches in ectodermal organoid cultures, we plan to dissect, how important these enzymes are for balancing the embryonic H4K20 methylation landscape, and how this is crucial for the successful differentiation of epidermal cells during development.
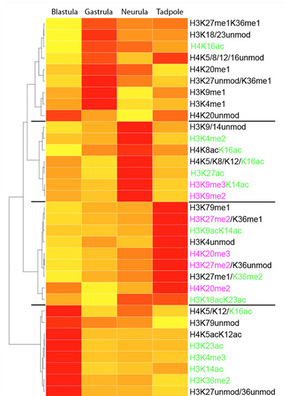
Figure 1: Heatmap detailing differential abundance of histone H3 and H4 modification states measured by tandem mass spectrometry (yellow-low, red-high abundance). The investigated developmental stages reflect key steps in embryonic development: Blastula – naïve multipotent; Gastrula – germ-layer specified; Neurula – germ layer patterning and differentiation; Tadpole – embryonic development completed.
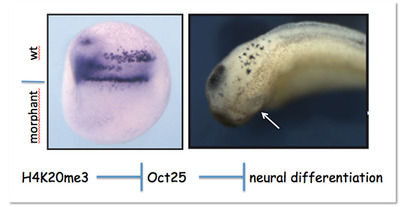
Figure 2: Downregulating H4K20 trimethylation by microinjection of Suv4-20h Morpholino oligonucleotides in one half of the embryo blocks primary neurogensis (left) and eye formation (right). This phenotype arises from perturbation of the regulatory pathway shown at the bottom.

Prof. Dr. Ralph RuppBiomedical Center - Molecular Biology, LMU Munich +49 (0)89 2180 -75438 or -75452 |

