A35 - Functional analysis and targeted manipulation of chromatin dynamics on mammalian noncoding regions
“Given the large amount of genome-wide data that have been collected during the last decades, a good understanding of how and why cells change during development, homeostasis, and disease might be expected. Unfortunately, the opposite is true; triggers that cause cellular state changes remain elusive, and the underlying molecular mechanisms are poorly understood. (…) Fortunately, recent methodological innovations are now providing options to answer these outstanding questions, by allowing to target and manipulate individual genomic and epigenomic loci. In particular, three experimental approaches are now feasible due to DNA targeting tools, namely, activation and/or repression of [genes] in their endogenous chromatin context; targeting transcription factors to endogenous, alternative, or inaccessible sites; and finally, functional manipulation of the chromatin context.” (Breunig et al., 2021).
In the suggested project, we will manipulate the expression of noncoding and RNA genes to test their relevance in the context of neural stem cell self-renewal and neuronal differentiation.
Breunig, C.T., Koferle, A., Neuner, A.M., Wiesbeck, M.F., Baumann, V., and Stricker, S.H. (2021). CRISPR Tools for Physiology and Cell State Changes: Potential of Transcriptional Engineering and Epigenome Editing. Physiol Rev 101, 177-211.
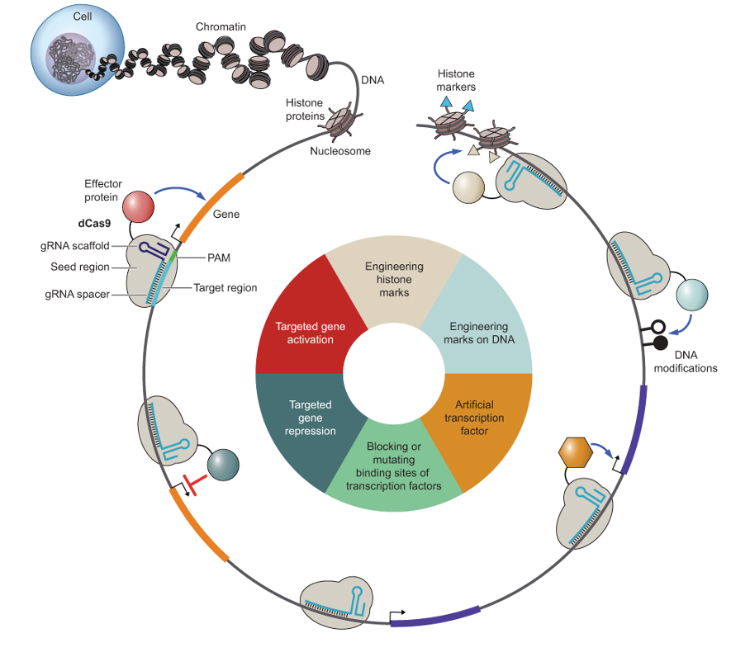
New molecular CRISPR tools to manipulate cellular states. The nucleus is the steering console ofthe cell. Molecular instructions for all possible cellular states are encoded in DNA. DNA is embedded in chromatin. CRISPR strategies include the activation and repression of critical genes, the interference with transcription factor binding, the use of artificial transcription factors, and the manipulation of chromatinfeatures. gRNA, guide RNA; PAM, protospacer adjacent motif. Figure and legend taken from (Breunig et al., 2021).


