A28 – Characterization of the chromatin organization response to food deprivation in the intestine of C. elegans
PhD position available in the Cabianca lab (posted 1 July 2022).
Multiple layers of regulation are required to maintain appropriate gene expression patterns. These include epigenetic mechanisms of non-DNA sequence-based chromatin modifications and higher order architecture of the genome. One hallmark of epigenetic regulation is that it is dynamic and can be altered by developmental and environmental cues. A major environmental input in biology is nutrient availability, with absence of food being a recurrent experience for animals and humans. Cellular metabolic pathways directly provide the donors for chromatin modifications, suggesting that the metabolic state of the cell can deeply influence the epigenome. However, it is largely unknown how altering the nutritional state of an organism impacts on the multiple layers of tissue-specific chromatin regulation and what implications this has for gene expression and organismal health.
Because of its rapid development, powerful genetics, robust response to dietary stress and the recent advances in tissue-specific tools, in this project, we will use C. elegans, and focus on food deprivation, also termed starvation, to study how metabolic stress affects tissue-specific chromatin regulation at multiple levels. In particular, we will measure i) modifications of histones, ii) spatial architecture of the genome and iii) gene expression within the same tissue of a multicellular organism in fed and starved conditions. Furthermore, the analysis of refed individuals will allow us to determine which starvation-driven alterations are restored and which are not, providing insights into epigenetic determinants of “stress memory”, a phenomenon that allows organisms to respond better to a second stress insult occurring later in life, in absence of the original one. Moreover, via a targeted microscopy screen, we will identify chromatin factors that functionally respond to food deprivation. Their genetic impairment will allow us to unravel mechanisms of chromatin re-organization under dietary stress and determine their role in gene expression and memory of the stress.
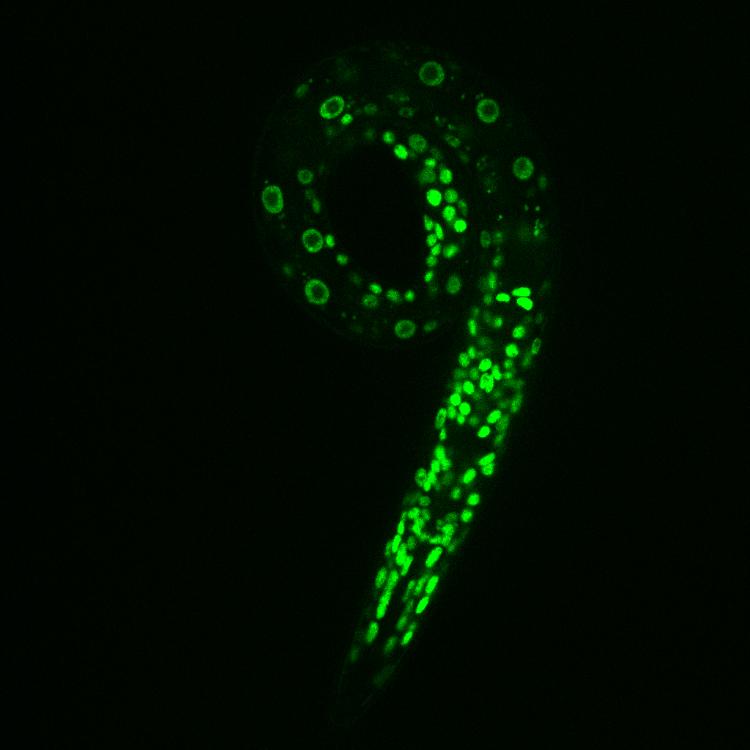
Live microscopy image of an early stage C. elegans larva expressing the histone variant H3.3 fused to GFP.
C. elegans is transparent throughout development, providing a unique opportunity to directly visualize fluorescently-labeled chromatin factors within intact tissues of a whole organism with live microscopy.


